Complimentary data analysis using our suite of bioinformatics tools
Free basic data analysis is included with all products and service orders through our iRWeb Bulk Repertoire and iPair Single Cell Analyzer systems.
Bulk Repertoire Analysis
iRweb
iRweb provides a comprehensive picture of immune repertoire diversity.
First, samples are demultiplexed and filtered to remove PCR and sequencing errors via our exclusive SMART sequence filtering system.
Then, reads are mapped to IMGT and all unique CDR3s are identified.
Our interactive interface helps you explore and compare the diversity of the repertoire for each chain in a BCR or TCR through proprietary visualization and diversity calculation tools.
Features
- Molecular ID/barcode sequencing decoding
- Downloadable demultiplexed raw or filtered data
- V, D, J, & C alignment to germline alleles
- CDR1, CDR2, and CDR3 identification (Long read kits)
- CDR3 identification (Short read kits)
- Identification of IgG, IgA, IgM, IgE, IgD, and subisotypes
- V-J combination distribution
- Identification and comparison of clonotypes
- Side by side repertoire comparison tools
- Diversity Index calculation
- Normalized and non-normalized distribution of N-addition, CDR3 length, V-J nibbling, and V-J usage
Interested in data analysis with iRweb?
Visualization tools
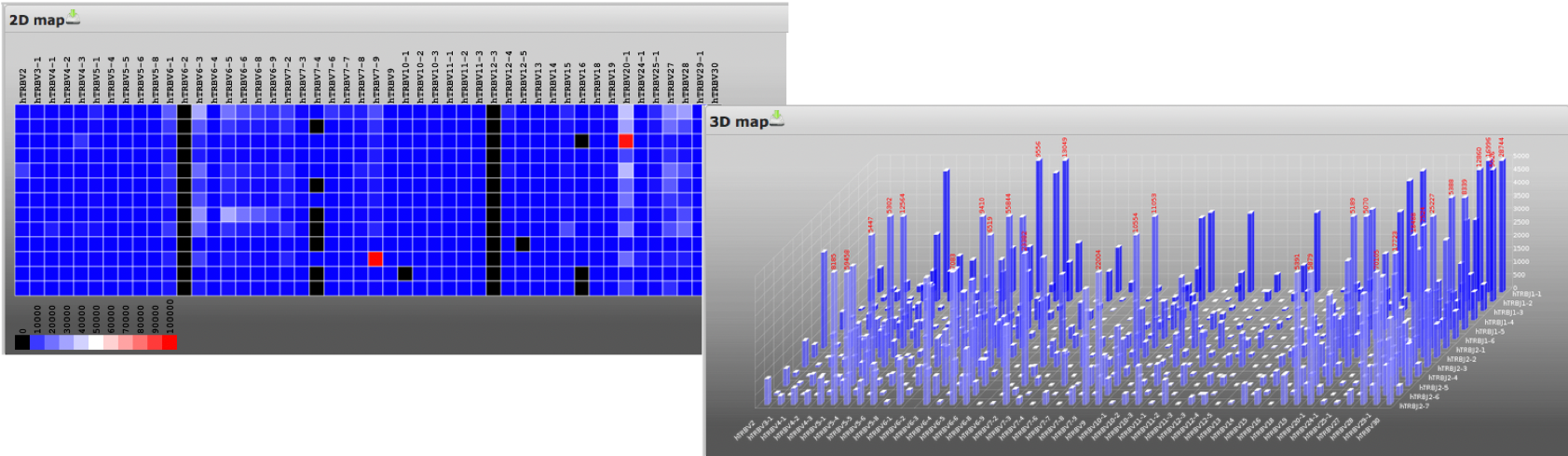
2D and 3D heatmaps
Heatmaps highlight the frequency of all identified V-J combinations within and between samples.
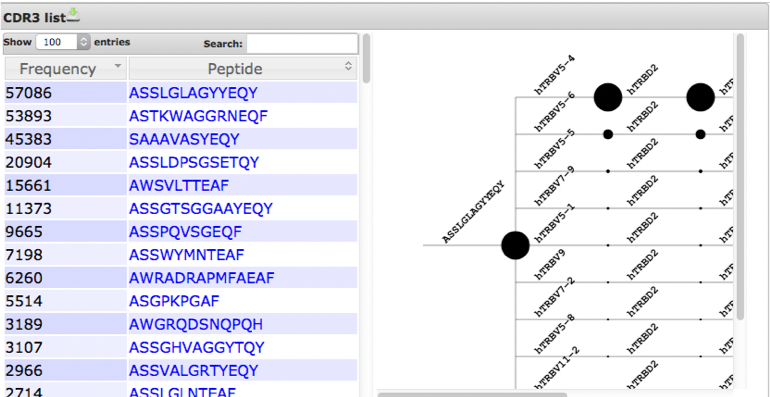
Hierarchical tree maps
Hierarchical trees break down all possible gene combinations for a given CDR3 peptide.
Diversity tree maps
Tree maps denote the relative frequency of unique V-J-uCDR3s as geometric shapes.

Single Cell Analysis
iPair Analyzer
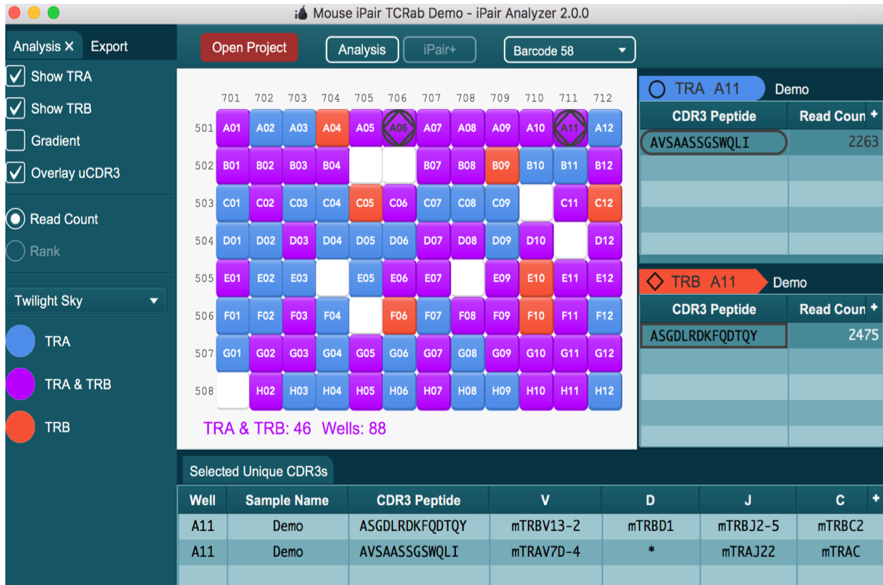
iPair Analyzer is an exclusive interactive tool that aids in visualization of TCR and BCR sequencing data for individual cells.
The iPair Analyzer graphic user interface represents samples in a 96 well plate via an interactive panel. When a cell in the panel is selected, single cell BCR and TCR chain results are displayed. A worksheet feature lets you select chains of interest and further explore and compare specific nucleotide sequences.
Hightable features
- Read depth
- unique CDR3s
- chain usage
- V/D/J usage
Phenotypic Analysis
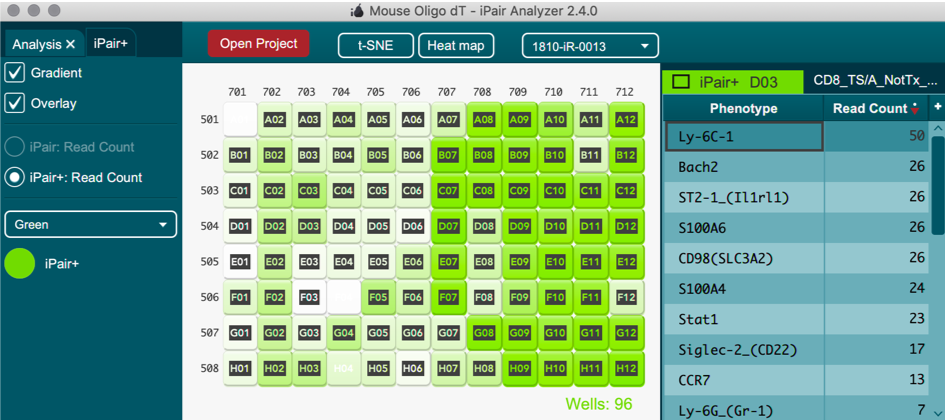
iPair+ Analyzer
With iPair+ single cell phenotyping data can also be visualized in iPair Analyzer via heatmaps and T-SNE plots.
When using iPair+, the Analyzer shows a gradient based on the number of reads for each well.