There are various sample collection methods researchers can utilize for immune repertoire studies. This document’s purpose is to define, compare, and contrast several methods of sample collection and downstream storage options that we currently use in-house as it applies to whole blood.
This document is not a comprehensive review of all possible collection methods but can be used by researchers to help determine the most appropriate collection strategy to achieve the desired results. We understand that samples are used for other applications in conjunction with immune repertoire studies; therefore, it is important to note that a sample can be stored multiple ways depending on the initial collection strategy. If other types of assays are planned, it might be necessary to collect multiple types of blood tubes at the time of collection.
Regardless of what path an individual researcher takes, we always recommend that cell samples used for immune repertoire analysis be stored using a nucleic acid stabilization media such as Qiagen’s RNAprotect Cell Reagent (cat. no. 76526). For tissue, we recommend RNAlater Stabilization Solution (cat. no. 76104). These reagents help to ensure high quality, intact nucleic acid following the extraction process. Other acceptable options for sample preservation include vacutainers containing RNA stabilization solution such as PaxGene tubes and cryopreserving cells (as long as correct freezing and storage procedures are followed).
For guidance on preparation of non-blood samples (isolated cells, tissues, extracted nucleic acids), see our sample submission guide.
Whole Blood: “Buffy Coat” versus PBMCs
Researchers generally collect whole blood as the starting material for immune repertoire analyses; however, researchers often then focus on specific cell subsets of interest. In this section, we will briefly review the makeup of whole blood and compare the definitions of “buffy coat” versus “PBMCs”.
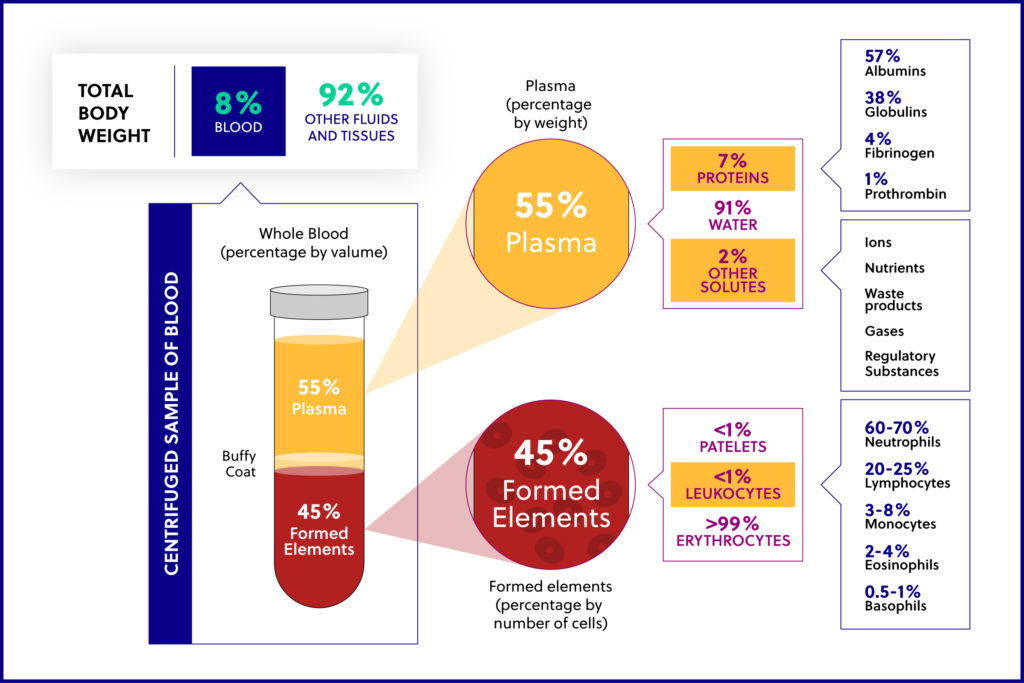
Whole blood contains red blood cells (RBCs), white blood cells (WBCs), platelets, and plasma. Plasma contains protein and additional nutrients. The WBCs include both peripheral blood mononuclear cells (PBMCs) and polymorphonuclear cells (PMNCs). Lymphocytes (T cells, B cells, NK cells), monocytes, and macrophages make up the PBMCs, while neutrophils, eosinophils, basophils, and mast cells make up PMNCs.
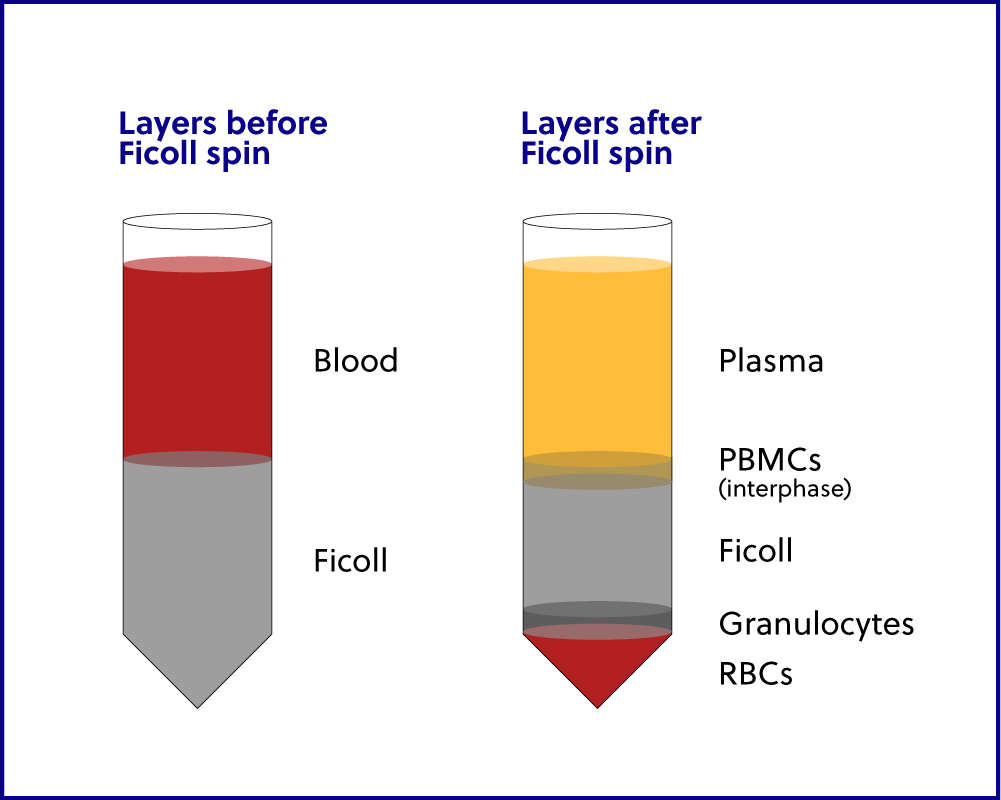
“Buffy coat” is the fraction of an anticoagulated blood sample that contains WBCs and platelets following direct centrifugation of whole blood. “PBMCs” are typically obtained by density gradient centrifugation using Ficoll or a similar reagent and include any peripheral blood cell with a round nucleus. While PBMCs are a type of WBC, they only account for a percentage of all WBCs. We use one method in-house to isolate the buffy coat from a whole blood sample, and we use a separate method to isolate PBMCs. It is important to note that the two terms defined here are NOT interchangeable.
Collection Methods for Isolating Buffy Coat and PBMCs
In this section, we will discuss why a researcher might want to use buffy coat or PBMCs and introduce a few collection methods that we use in-house to isolate the two types of samples from whole blood.
Total WBC
When the cells of interest for a particular study include cell subsets from both PBMCs and PMNCs, the buffy coat should be isolated from whole blood. This can be achieved by direct centrifugation of whole blood (refer to online sources for specific methods related to buffy coat isolation) or through RBC lysis. We typically, at minimum, extract from 10 mL of whole blood for immune repertoire studies. After the buffy coat is isolated, it can be processed for several downstream applications: nucleic acid extraction, cryopreservation, storage on Qiagen’s RNAprotect Cell Reagent (cat. no. 76526), or perhaps a combination of these three approaches depending on suspected downstream usage, if desired.
PBMC Isolation
When the cells of interest do NOT include cells from both PBMCs
and PMNCs, then we recommend using a Ficoll gradient method to separate the
WBCs into distinct layers, as illustrated in Figure C and discussed below.
Isolation of PBMCs is usually recommended for any sort of downstream cell
sorting. Ficoll separation can be used for most whole blood samples drawn into
a vacutainer with an anticoagulant additive. Our preferred vacutainer is the
BD K2-EDTA blood collection tube. Following dilution, layering over Ficoll
and centrifugation, the PBMC layer is easily isolated and can be processed for
several downstream applications: nucleic acid extraction, cryopreservation,
cell sorting (bench-top or FACS), storage on RNAprotect Cell Reagent, or
perhaps a combination of these four approaches if necessary.
The volume of blood drawn depends upon the
downstream process. If RNA will be extracted directly, 10 mL is recommended as
a minimum. For sorting cells, the frequency of the downstream cell subset
affects this decision. In-house, for some studies, this can vary from 20-40 mL
of whole blood. Please see iRepertoire’s Sorting&RNAExtracting Guide for
more information.
Ficoll gradient separation can be used to isolate a distinct layer
of whole blood, most often PBMCs, and can be performed using two different
types of vacutainers.
- To perform manual Ficoll gradient separation, whole blood may be
drawn into a vacutainer with an anticoagulant additive. Again, our preferred
vacutainer is a BD K2-EDTA blood collection tube. Following dilution with a
specific buffer, the diluted blood is layered over a commercially available Ficoll
gradient solution and then centrifuged as detailed in iRepertoire’s
Sorting&RNAExtracting Guide. Note: heparin additives in blood collection
tubes can inhibit downstream reverse transcription and PCR if certain nucleic
acid isolation techniques are used.
- To perform Ficoll gradient separation directly from the whole blood, whole blood may be drawn into a vacutainer with a Ficoll additive. Our preferred vacutainer is the a BD Vacutainer CPT cell preparation tube with sodium citrate. Following the blood draw, the tube can be centrifuged and PBMCs decanted from the tube.
Comparison of different collection tubes
In this section we will list the advantages of drawing blood into either the BD K2-EDTA blood collection tube or the BD Vacutainer CPT cell preparation tube with sodium citrate.
BD K2-EDTA Blood Collection Tubes
Advantages:
- Stored at room temperature until processing
- Can isolate either the buffy coat or PBMCs depending on how the blood is processed
- A total WBC count be taken directly from the blood tube
- Has a blood anticoagulant
- EDTA is compatible with downstream RT-PCR applications (unlike sodium heparin which can inhibit RT enzymes)
- Available in multiple sizes depending on how much blood is needed (2, 3, 4, 6, and 10 mL options)
- Provides the most options for processing: RBC lysis, buffy coat isolation, and Ficoll gradient separation
Limitations:
- Must be processed within 24-48 hours from collection
- Requires storage at room temperature on a rotator to prevent cell death and partial coagulation
- Does not contain any nucleic acid stabilization additive
- Requires manual processing, which can be time consuming
BD Vacutainer CPT Cell Preparation Tube with Sodium Citrate
Advantages:
- Stored at room temperature until processing
- Time efficient way to isolate PBMCs (no need to manually add a Ficoll gradient solution; brake can be used during centrifugation, which greatly reduces processing time)
- Reduces consumables waste in the lab
- Can be drawn alongside other blood collection tubes
Limitations:
- Must be processed within 2 hours from collection
- The PMNCs cannot be easily isolated once blood is drawn into the tube (a protocol is provided by the manufacturer but the layer is not easily accessible)
- Higher initial cost than the BD K2-EDTA Blood Collection Tube
- Does not contain any nucleic acid stabilization additive
- Limited to 4 or 8 mL per tube
PAXgene Blood RNA Tubes
In this section we will discuss why a researcher might want to
consider a third type of blood collection tube, the PreAnalytiX PAXgene blood
RNA tube. We will also list its advantages and limitations.
When the cells of interest for a particular study include all cell
subsets from both PBMCs and PMNCs, and total RNA from WBCs is the intended
nucleic acid for downstream applications, the whole blood could be drawn
directly into a vacutainer designed to immediately stabilize the RNA. Our
preferred vacutainer is the PreAnalytiX PAXgene blood RNA tube. Following the
blood draw, the tube can be centrifuged and processed using the PAXgene Blood
RNA Kit for the extraction of high quality, intact RNA.
Advantages:
- Can be stored at room temperature for up to 3 days
- Can be stored at 2-8°C for up to 5 days
- Can be stored at -20°C or -70°C for up to 11 years
- Contains RNA stabilization additive
- Time efficient way to isolate total RNA from whole blood (no RBC lysis processing)
- Can be drawn alongside other blood collection tubes
- Can be thawed at room temperature
- Processed using the corresponding PAXgene Blood RNA Kit
- No need to process the tube until the RNA is needed
- Often used in clinical studies
Limitations:
- Requires an incubation period of 2 hours prior to processes
- Cell subsets (PBMCs and PMNCs) cannot be isolated separately once blood is drawn into the tube
- Higher initial cost when using the blood collection tubes and corresponding extraction kit
- Not compatible with cryopreservation, cell sorting (bench-top or FACS), or storage of specific cell subsets on RNAprotect Cell Reagent
- Limited to 2.5 mL of whole blood per collection tube
- Special instructions provided by the manufacturer for the blood draw (phlebotomist needs to familiarize themselves with the process prior to beginning)
RNA extracted from blood
For service samples, we request more than is
required for a single reaction so that there is enough for a repeat
amplification should it be necessary. The amount of RNA input is dependent
upon the starting sample type. If you are using total RNA from whole blood (no
sorted cells), we recommend using 1,000 ng of RNA input. This is the same
recommendation for tumor or various tissue types. While this input might seem
high, keep in mind that non-immune specific RNA is also in the sample. This RNA
will act as background noise, so we want to ensure that enough immune-specific
RNA goes into the reaction.
As the sample gets more immune-specific (for example, purification of PBMCs from whole blood enriches the present adaptive immune system), the suggested RNA input goes down. Service projects using PBMC-specific RNA averages anywhere from 300-500 ng input given sample availability. The suggested input goes down even more when a sample contains sorted cells. This is because the concentration of immune-specific RNA is greater. When working with sorted cells, we have used as little as 20 pg (good quality, intact) RNA. When considering how much RNA to input, always take into account the source of the RNA and the RNA quality. RNA quality is typically more important than quantity.
For guidance on collection/handling/submission of all other types of samples and for general tips on working with RNA, please see our sample submission guide