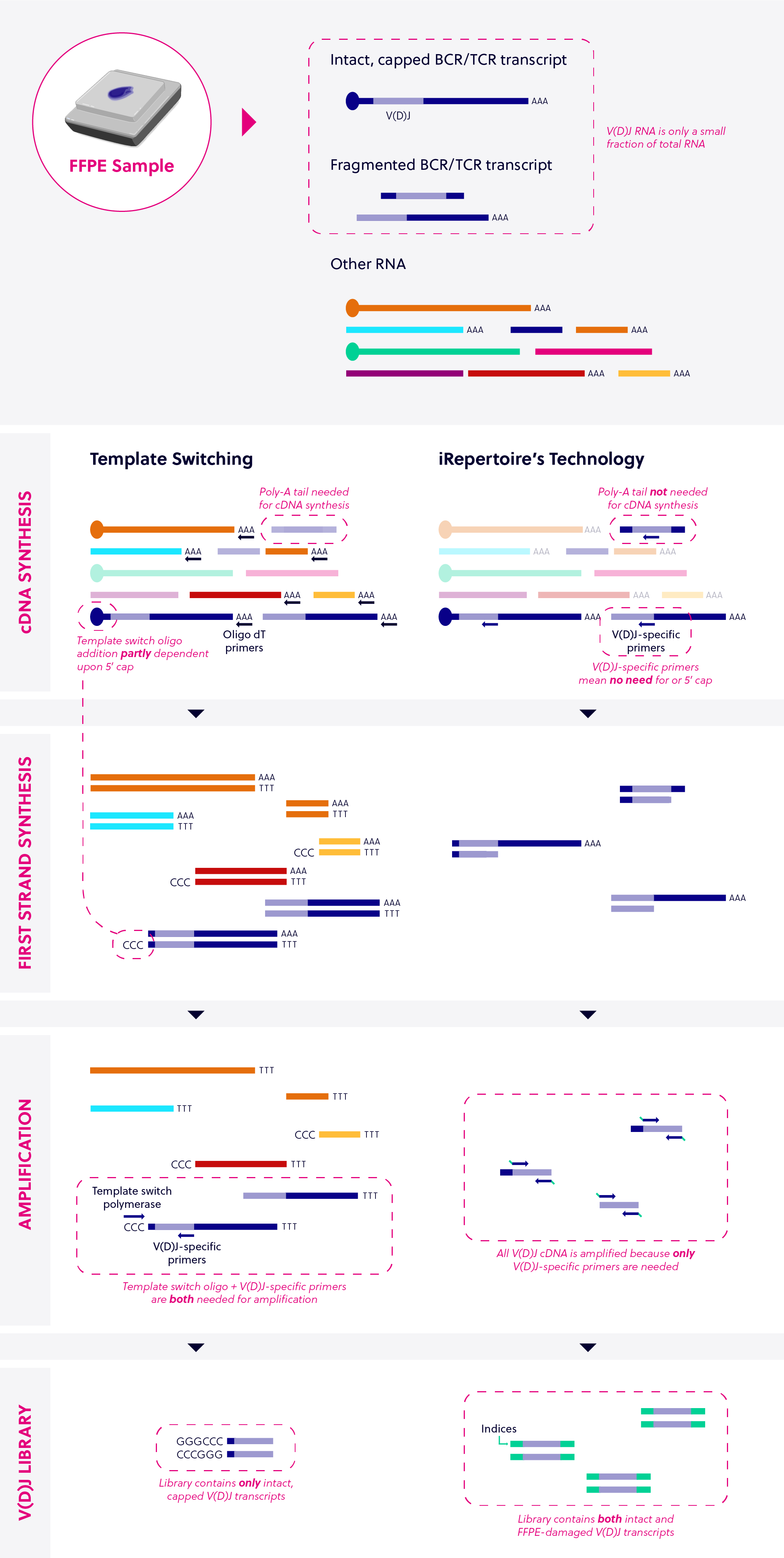
Investigating the full clonal diversity of an immune repertoire requires a uniquely tailored sequencing approach. Most immune sequencing chemistries on the market rely on some variation of a technology called template switching, which starts with untargeted amplification of all of the RNA in a given sample. iRepertoire’s proprietary multiplex PCR technologies use primers that are specifically designed for immune sequencing, enabling more sensitive and quantitative amplification.
What is template switching?
The benefit of template switching – in theory – is that it allows for the capture of full cDNA sequences, for which the 5’ end is unknown, in a relatively sensitive manner. Therefore, variations of template switching are often used in shotgun sequencing of the whole transcriptome.
The first step is to capture virtually any mRNA with a poly A-tail present in the sample by performing reverse transcription with a 3’ oligo-dT primer. What distinguishes template switching from other random poly-A tail amplification is a secondary function of the reverse transcriptase that incorporates a string of non-templated base pairs at the 5’ end of the cDNA under certain conditions.
The non-templated bases are then targeted with a specifically designed 5’ oligo. The added bases become the template (hence the name, template switch). In this second amplification step, 3’ oligos specific to the target(s) of interest can be used to pull out a particular population of RNA; in the case of immune sequencing, B or T cell receptor genes.
How is iRepertoire’s technology different?
iRepertoire’s multiplex PCR technology targets V(D)J RNA from the start, providing more sensitive and robust amplification of immune sequencing targets. The immune repertoire of any given sample will be composed of some clonotypes that are exceedingly rare and some that are overwhelmingly abundant. While template switching can be used to specifically amplify BCR and/or TCR genes, during the first step of template switching, mRNA is converted to cDNA indiscriminately. This is reflected in the end sequencing results: many non-VDJ RNA will be included, reducing the desired signal. Additionally, the non-templated base pair incorporation step is biased toward capped RNA structures, so the quality of the sample is paramount.
Advantages of iRepertoire’s technology over traditional template switching:
- Detection of extremely rare clonotypes is possible
- Robust to RNA degradation allowing difficult sample amplification
- Selective chemistry options to meet specific project goals
- Unmatched sensitivity and bias reduction
iRepertoire’s multiplex PCR technologies specifically reverse transcribe and amplify BCR and TCR transcripts right from the first step, which enables the detection of extremely rare clonotypes and maintains the relative abundance of different clonotypes. Because a portion of the receptor can be targeted, iRepertoire’s technologies are also more robust to RNA degradation, enabling amplification of difficult samples such as FFPE RNA. Depending on the goals of your project, you can choose from two different chemistries specifically developed for immune repertoire sequencing.
| Template switching | iRepertoire’s Technology |
|---|---|
| Poly-A tail needed for cDNA synthesis | Poly-A tail not needed for cDNA synthesis |
| Template switch oligo addition partly dependent on 5’cap | V(D)J- specific primers amplify independent of the 5’cap |
| Library contains only intact V(D)J transcripts and off-target expression products | Library contains both intact and FFPE-damaged V(D)J transcripts |
Arm-PCR
Arm-PCR, iRepertoire’s flagship technology, is catered towards rare clonotype discovery and enables a semi-quantitative comparison of relative clonotype frequency. The extreme sensitivity of arm-PCR is accomplished using a nested series of inside and outside primers that recognize the V and C region of B and T cell receptor transcripts. The outside primers increase the target pool while the inside primers append communal adaptors. In a second series of amplification steps, primers targeting the communal adapters are used to increase the signal, ensuring that only full length, spliced VDJC’s proceed to sequencing.

Dam-PCR
Dam-PCR is exceptionally quantitative and optimized for error and bias reduction. Dam-PCR starts with a single-cycle binding and extension step using template-specific primers. A clean-up step occurs next to eliminate unused primers. This step, along with the addition of unique molecular identifiers (UMIs), makes dam-PCR the most quantitative immune sequencing technology on the market. The presence of UMIs also assists with identification of PCR and/or sequencing errors during bioinformatic analysis. The ability to accurately identify PCR and/or sequencing error is especially beneficial to BCR repertoire analysis where somatic mutations are abundant.

For both of iRepertoire’s proprietary multiplex PCR solutions – arm-PCR and dam-PCR – the steps for isolating the target RNA and the steps for exponentially amplifying those sequences are physically and temporally separated, which enables unmatched sensitivity and bias reduction.