iRepertoire’s iR-Complete Dual Index Kits will replace both the iR-Profile and Bulk Primer kits to simplify reagent ordering and promote scalability of your immune sequencing experiments.
Dual index adaptors for improved scalability
The previous iR-Profile kit contained 10 uniquely-barcoded reactions with primers tailored to the species and chains of the user’s choice, along with reagents for PCR such as amplification, rescue, secondary amplification and cleanup. In order to scale immune repertoire sequencing projects, Bulk Primer kits were required. Bulk Primer kits contained internally barcoded primer mixes with communal adaptors for universal PCR2 amplification. Although each bulk primer kit contained enough primers to make 5 or 10 libraries, these libraries were all of 1 internal barcode. For increased multiplexing, purchase of multiple kits was previously required in order to fully take advantage of the read depth of a variety of Illumina flow cells.
A new approach to sample barcoding is at the core of our iR-Complete Dual Index Primer Kits. Scalable multiplexing is now possible with the use of a single kit.
A single kit for simplified library preparation
The iR-Complete Dual Index Primer Kit for RNA or gDNA utilizes the same targeted primer selection, as previously available, for amplifying the highly variable CDR3 receptor region, critical to defining the immune repertoire. Kits include reagents for generating sequencing-ready libraries. Detailed procedures are available. In addition, RNA-based iR-Complete kits include a proprietary wash buffer and procedure for difficult to amplify samples such as FFPE, providing data for precious and previously unyielding samples.
Just like our legacy products, an internal sample barcode is incorporated into the amplicon during PCR1 followed by the addition of dual index sequencing adaptors in PCR2. The combination of internal and dual index adaptor generates a unique labeling system for each sample. Libraries are sequencing-ready and can be sequenced on any Illumina platform including iSeq®, MiniSeq®, MiSeq®, NextSeq®, and NovaSeq®. For existing customers, it is important to note that the PCR2 dual index adaptors available in iR-Complete kits are not compatible with legacy iR-Profile or Bulk Primer mixes.
With unique combinations of internal barcodes and dual index adaptors, up to 96 individual libraries can pooled, sequenced, and analyzed simultaneously. Universal Dual Index Kits are offered in as 12-, 24-, 48- or 96 reaction kits to accommodate a range of customer throughput requirements (Figures 1-4):
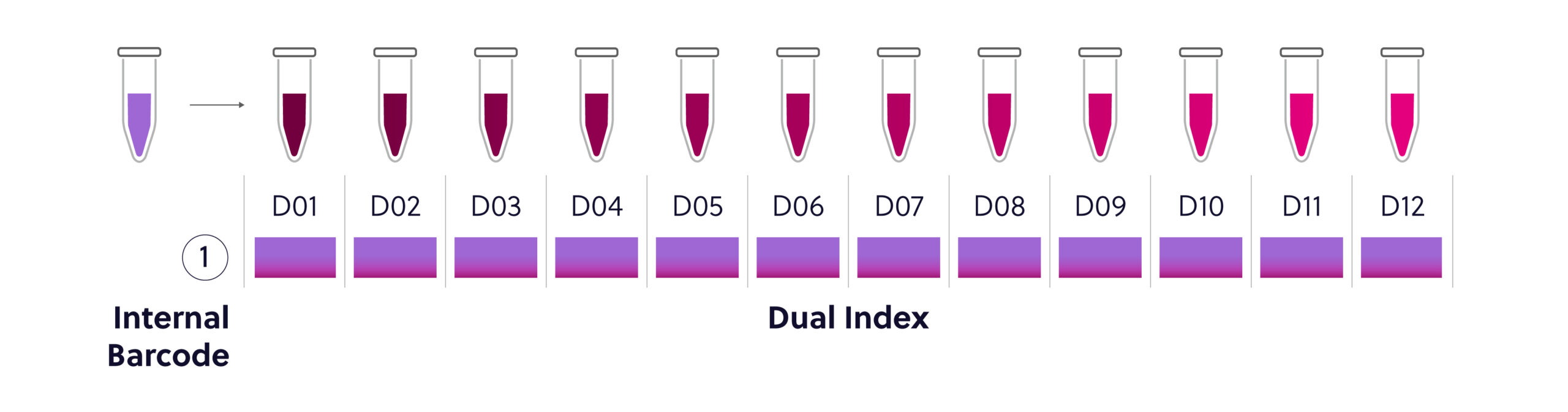
Figure 1. 12 reaction Universal Dual Index Kit configuration. This kit contains 1 internal barcode and 12 individual dual index adaptors. This is the simplest configuration of the kit and can create 12 uniquely barcoded libraries.
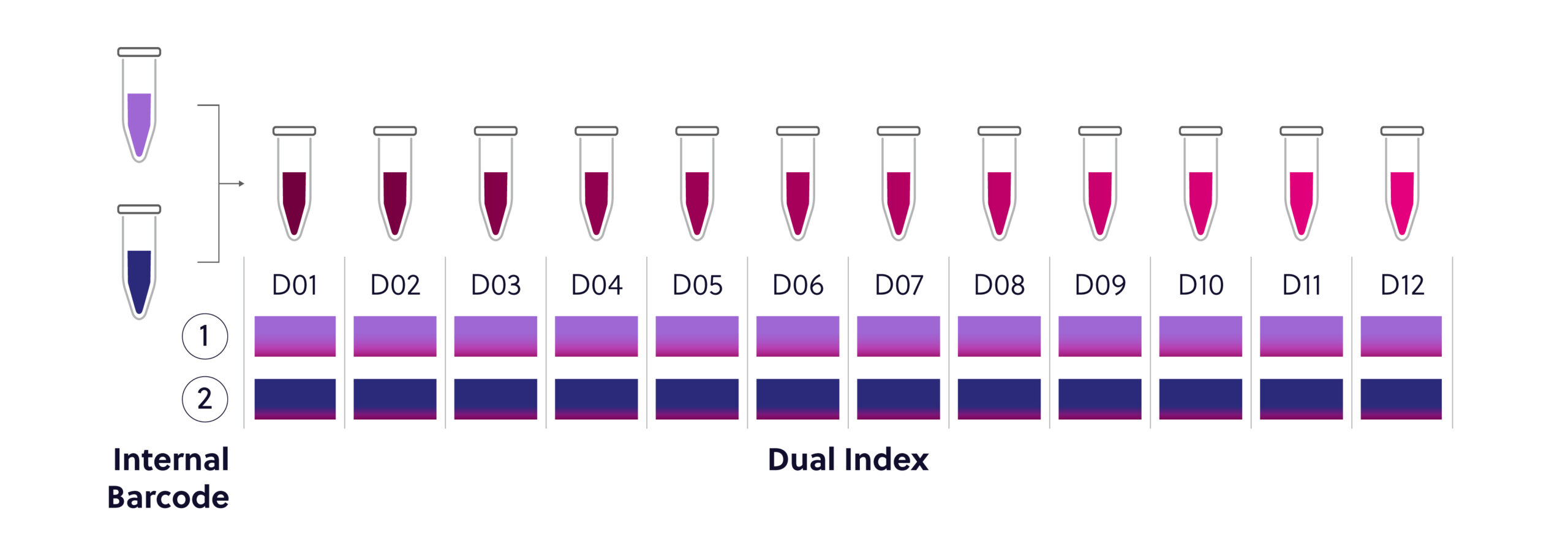
Figure 2. 24 reaction Universal Dual Index Kit configuration. This kit contains 2 internal barcodes and 12 individual dual index adaptors. Ensure that a unique combination of internal and external barcodes is utilized for each sample.
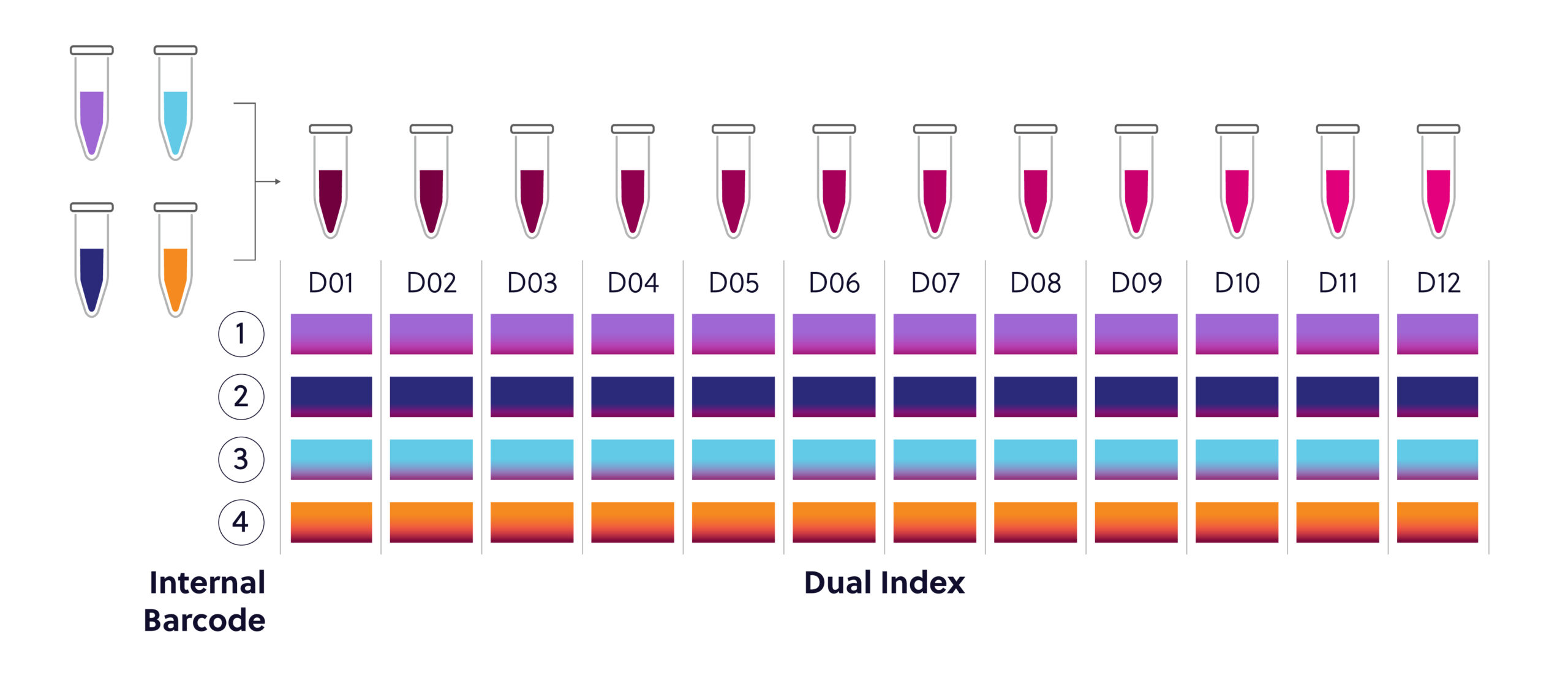
Figure 3. 48 reaction Universal Dual Index Kit configuration. This kit contains 4 internal barcodes and 12 individual dual index adaptors. Ensure that a unique combination of internal and external barcodes is utilized for each sample.
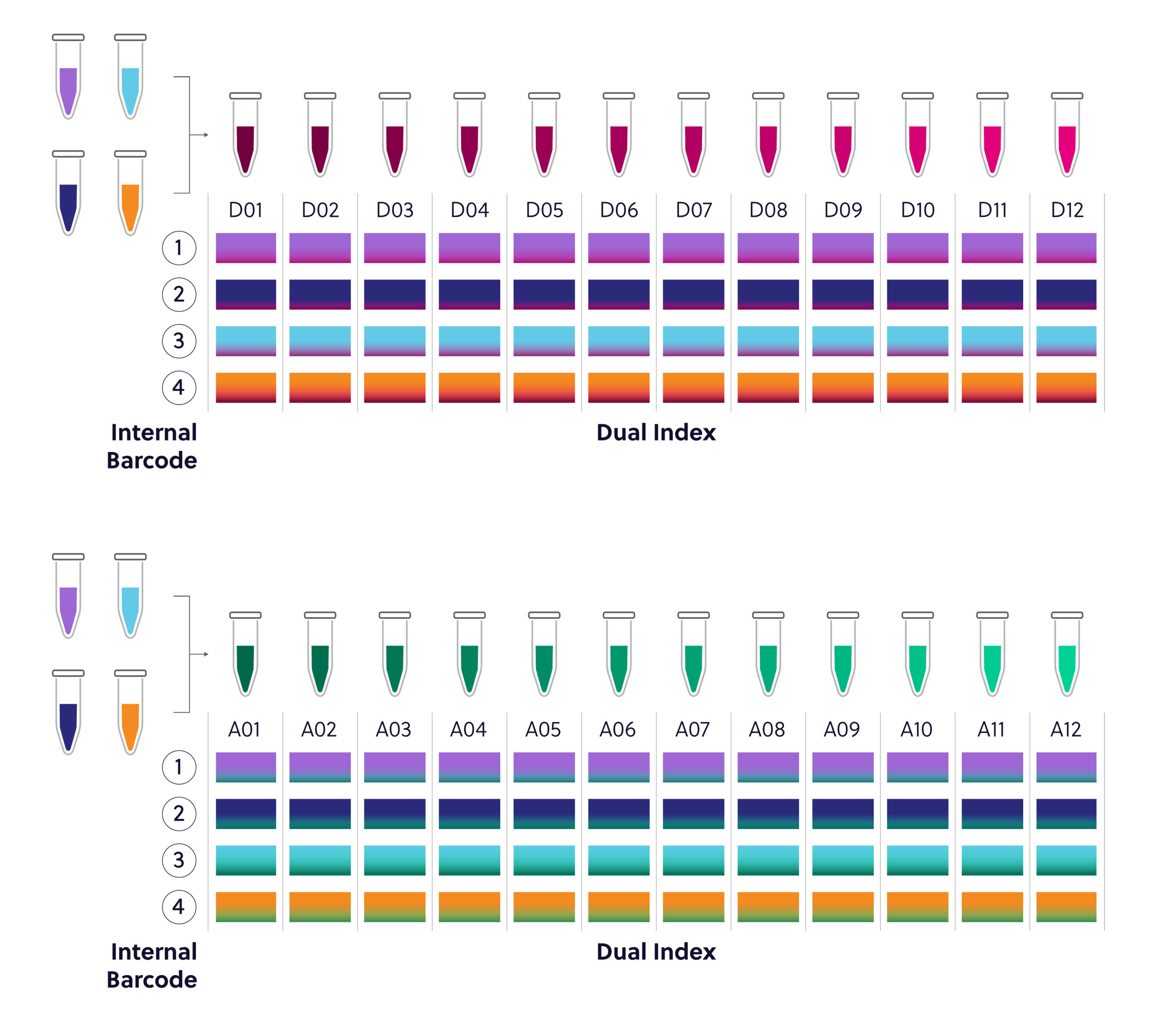
Figure 4. 96 reaction Universal Dual Index Kit configuration. This kit contains 4 internal barcodes and 24 individual dual index adaptors. Ensure that a unique combination of internal and external barcodes is utilized for each sample.
Next-Level Immune Sequencing
The Universal Dual Index Kit is designed for user simplicity and to support immune sequencing scalability. Kits are tailored for a range of throughput requirements that enable customers to control costs, reduce reagent waste, and support any immune sequencing application.
Is the data comparable between legacy products and the new dual index kits?
Extensive testing with the dual index primer sets has been performed at iRepertoire indicating that there is no major difference between the libraries generated with the dual-indexed versus the non-indexed primer sets. The primary distinction between iR-Complete Dual Index Primer kits and legacy Bulk Primer and iR-Profile kits is the inclusion of dual indices during PCR2. The PCR1 primers and PCR1 thermal cycler conditions are the same, and thus the endpoint library data are very similar. Below are some figures indicating the similarities between data produced by dual-indexed and non-indexed libraries for HTAI-M and HTBI-M primer sets. The dual-indexed and non-indexed repertoire results are first compared in terms of reads, unique clonotype discovery, and similarity of diversity metrics such as D50, Diversity Index, and Shannon Entropy. Next, three overall comparisons are done to evaluate repertoire differences: 1) the repeatability of shared clonotypes and their frequency across replicates (R2 of shared clonotypes) is observed 2) the shared expression (shared clonotypes and their associated reads) across replicates is calculated and 3) the total number of shared clonotypes across replicates (without considering frequency) is observed. All of these metrics fall within established, acceptable QC ranges. Within the dual index data, comparisons of repeatability can also be made across different internal and dual index barcodes. These data show an expected level of consistency across the replicates regardless of internal and external sample barcoding.
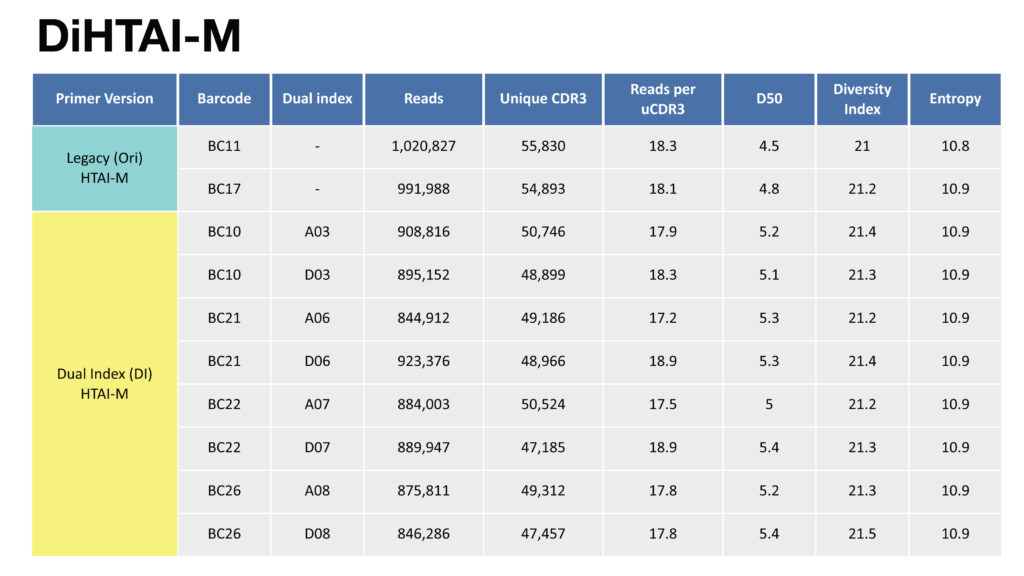
A. For primer panel HTAI-M, comparison table of reads, uCDR3, and various diversity measurements between non-indexed legacy primer sets and dual indexed primer sets.
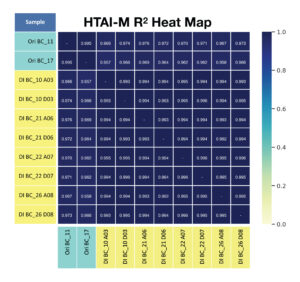
B. For primer panel HTAI-M, the repeatability of shared clonotypes and their frequency across replicates (R2 of shared clonotypes) is provided.
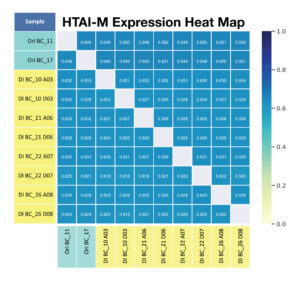
C. For primer panel HTAI-M ,the shared expression (shared clonotypes and their associated reads) across replicates is calculated.
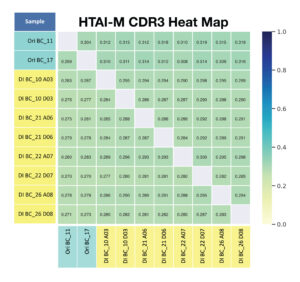
D. For primer panel HTAI-M, the total number of shared clonotypes across replicates (without considering frequency) is observed.
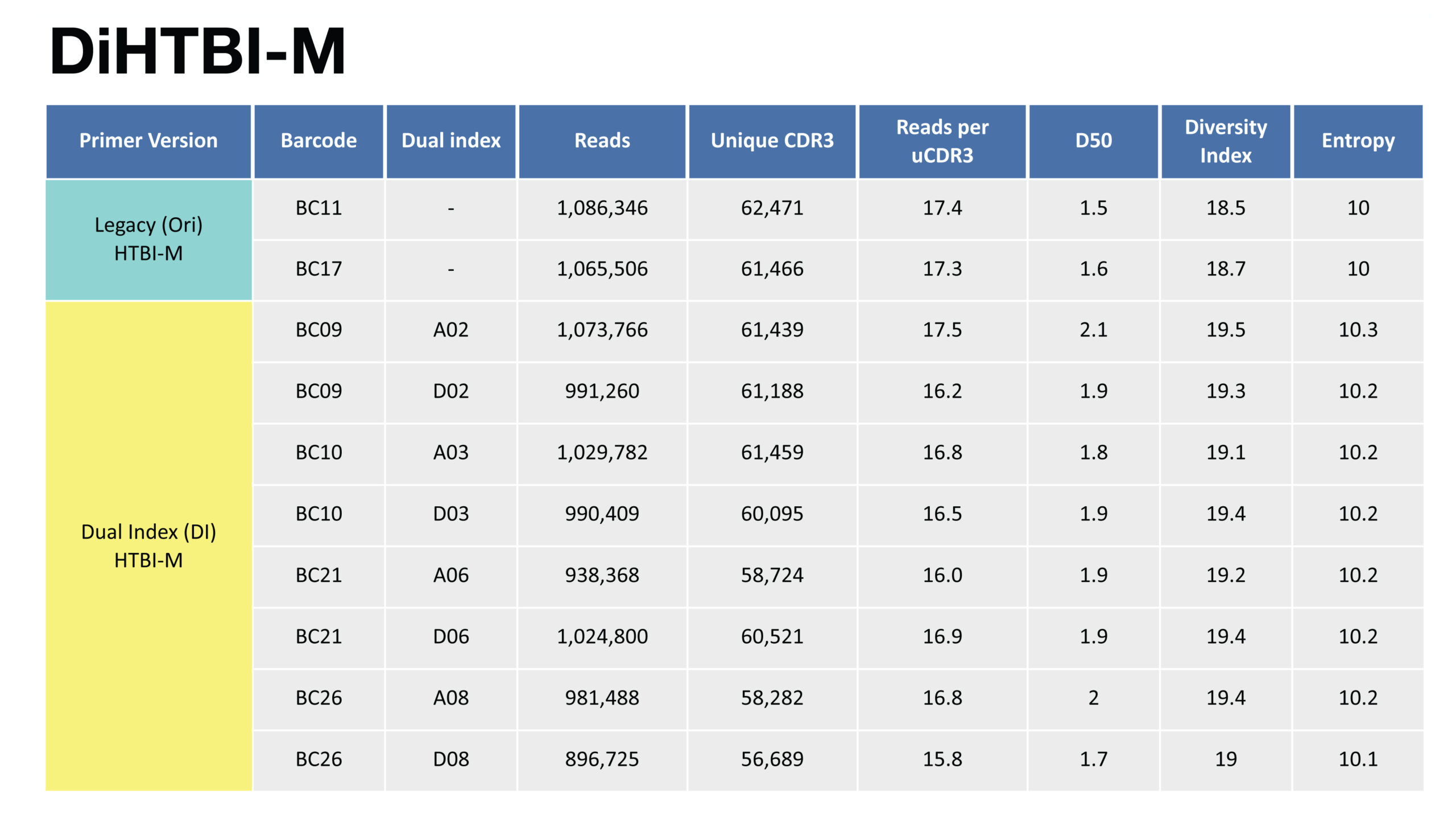
A. For primer panel HTBI-M, comparison table of reads, uCDR3, and various diversity measurements between non-indexed legacy primer sets and dual indexed primer sets.
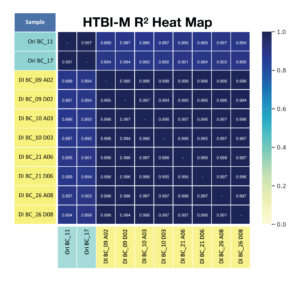
B. For primer panel HTBI-M, the repeatability of shared clonotypes and their frequency across replicates (R2 of shared clonotypes) is provided.
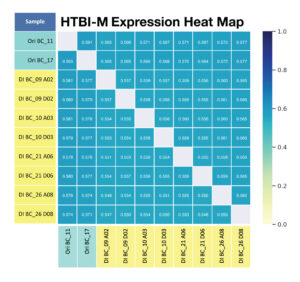
C. For primer panel HTBI-M ,the shared expression (shared clonotypes and their associated reads) across replicates is calculated.
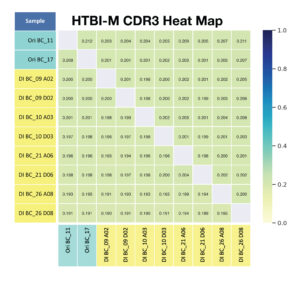
D. For primer panel HTBI-M, the total number of shared clonotypes across replicates (without considering frequency) is observed.
For more information about how to order kits for your application, contact us.