Why single cells?
Information related to the physical, cognate pairing of TCR-alpha and -beta chains or Ig-heavy and -light chains is lost once RNA extraction is performed on a bulk cell sample. iPair, our single cell immune sequencing service, was developed so that the repertoire of individual cells could be profiled through capture of chain pairing without the need to culture cells.
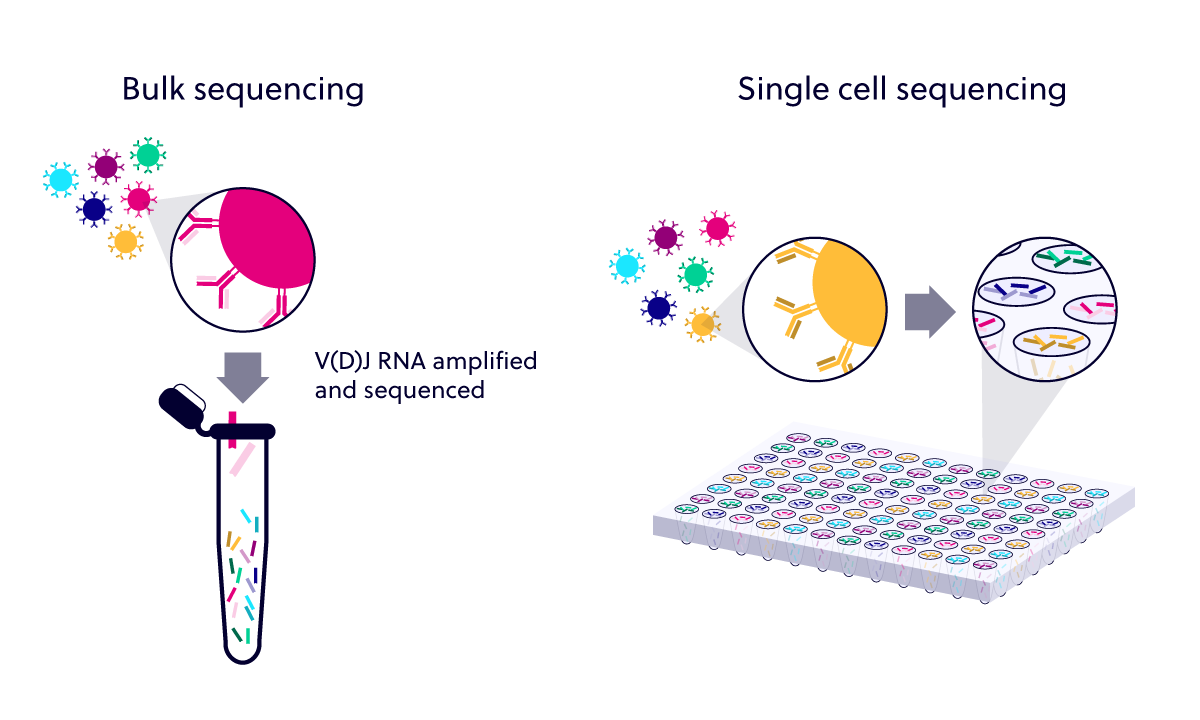
iPair was introduced on the market in November 2016. To our knowledge, iPair was the first consumer-based service specifically designed and intended for capturing the physically paired TCR alpha/beta or BCR IgH/KL variable region from human T and B cells. Since then, our services have expanded to cover mouse samples, as well as development of TCR gamma/delta pairing.
How it works
iPair relies on our arm-PCR technology. With iPair, we use a FACS-based approach to plate single cells into a 96-well iCapture plate. Our multiplex primer mixes were developed to balance the amplification of both loci simultaneously in the same PCR well in a low reaction volume. We then sequence the results using the Illumina MiSeq platform with 500+ cycle sequencing kits so that the variable region sequence can be called.
iPair generally achieves a >90% amplification success rate, and the pairing success rate is greater than 80% with our all-in-one targeted-seq mixes.
iPair primers
The iPair primers were designed using our long-read primer system, which covers from just within FR1 to the beginning of the C-gene for human samples (mouse long-read primers begin within FR2). Since we are only missing the first ~30-50 nucleotides of the V-gene, these sequences can be inferred from the reference sequences on the international ImMunoGeneTics information system (IMGT) quite easily. The placement of the primers produces libraries that are ideally sized for 500-cycle kits on Illumina platforms (250 paired-end reads). We have had many clients reconstitute functional TCRs from the data using the inferred sequences without issue as the majority of the TCR is present in the data including CDR1, CDR2, and CDR3.
To learn more about iRepertoire’s primer systems, please see our Learning Center article on iRepertoire’s primer system.
Single cell phenotyping with iPair+
Our single cell phenotyping service, iPair+, provides additional functional information about the cell(s) profiled using specialized primers. There are two different customizable options for iPair+, and which is best for your project largely depends on which phenotypic targets you’d like to call, how many targets you have, and your budget.
The gene-specific primer method involves co-amplification of phenotypic targets and the VDJ receptor primers in the same well. There are 30 gene-specific primers available for human samples; mouse samples must be processed with the oligo-dT approach.
The main advantage of using gene-specific primers is affordability. Because everything is processed in a single plate, there are no extra processing fees. However, highly expressed phenotypic targets could occupy a large amount of reads during sequencing, which could cause VDJ dropout. We recommend using the gene-specific approach if only a low number (~10-15) of phenotypic markers will be used.
Our second approach is an oligo-dT based method. The oligo-dT panel includes 100+ gene targets for either human or mouse samples. During this process, the cells are lysed and RT is performed with an oligo-dT primer mix. This cDNA is then split between two plates: one for VDJ amplification, and the other for phenotyping. Because the cell in each well is individually barcoded, phenotypic data can be overlaid with VDJ information during downstream data analysis. While this does increase the processing price, it allows for more user control of the sequencing depth of phenotypic targets.
Please contact us to view the gene-specific or oligo-dT panel lists.
Jump to iPair+ page for specific information
Applications
New applications in immunotherapy and an increasing awareness of the importance of the tumor microenvironment have produced a growing demand for immune profiling at the single cell level. iPair can be used in CAR-T development, for calculating clonal frequency in cell subsets, or to track specific lymphocytes following treatment.
Contact us to learn more about how you can use iPair.