iRepertoire has developed multiplex PCR primer mixes for V(D)J amplification and sequencing for both our arm-PCR and dam-PCR technologies. In these multiplex PCR systems, a nested set of inside and outside primers has been designed and optimized for every V(D)J target needing to be co-amplified in the multiplex assay. We offer primers, amplification services, and sequencing services for all TCR and BCR chains for both human and mouse species in order to meet your research needs, whatever they may be.
For more information about arm-PCR and dam-PCR, see our amplification technology page.
iRepertoire offers several different primer systems
iRepertoire has multiple different primer systems that vary by the regions targeted, the desired read length, and the species. Regardless of what primer system works best for your study, all of our primers cover the highly variable CDR3 region at a minimum, as this is generally the greatest area of diversity and interest.
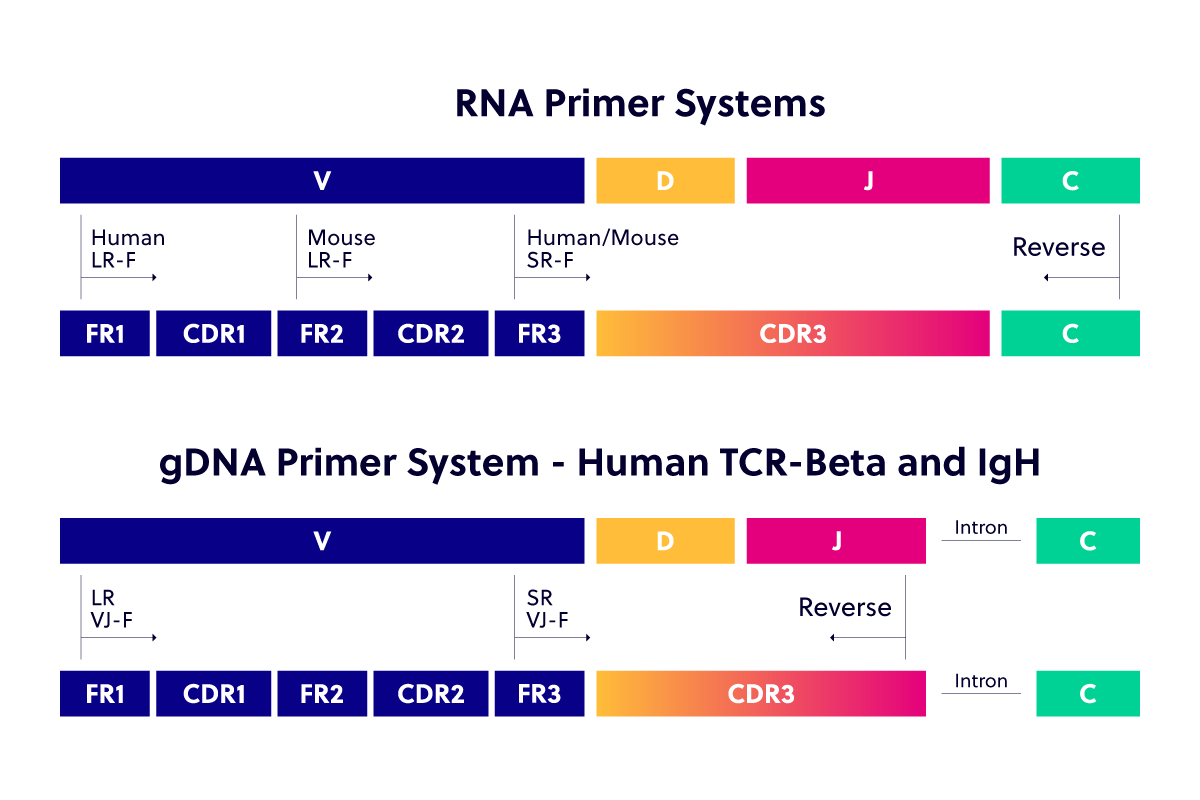
Our primer technology essentially grew alongside sequencing and read depth ability. Our services began with our short-read (SR) RNA-compatible human and mouse primers, which cover from within Framework-3 (FR3) into the Constant Region (C). These SR primers are compatible with 100/150 paired end read (PER) sequencing. As sequencing technology became more advanced, our long-read (LR) primer systems became available. These cover from within FR1 for human samples and FR2 for mouse, and both continue through to the C region. iRepertoire’s LR primers are compatible with 250 PER sequencing.
While we typically prefer RNA as the starting template (see our LC article on choice of starting template), we understand some researchers’ preference for gDNA. Due to a large intron gap between gDNA VDJ and C domains, it is necessary to use a different primer set to cover gDNA. Therefore, we have our V-J primer system for use with gDNA as the starting template (available for Human TCR-Beta and Human IgH).
The original gDNA compatible primers (SR-VJ) covered from within FR3 to the end of the J-gene. These have been replaced with our newest dual index and gDNA compatible VJ offerings (LR-VJ), which cover from within FR1 to the end of the J-gene. LR versions are available for both human TCR beta and human IgH. These can be sequenced as single end read on 300-cycle kits or for full amplicon coverage as 250 PER sequencing.
To learn more about PER and sequencing depth, see our guide on choosing read length/depth.