Challenges of immune repertoire sequencing
The unique challenges involved with sequencing the immune repertoire have necessitated the development of repertoire-specific library preparation technologies. In order to fully capture the diversity of the immune repertoire, amplification technologies have to be used that account for biases towards small amplicons and amplifying rare clonotypes beyond the templates that are naturally high in abundance. This is no small feat considering there are multiple chain types that contribute to each individual clonotype in a given repertoire, and the diversity of the repertoire has to account for gene recombination, expression, and hypermutation.
iRepertoire’s multiplex PCR amplification technologies were specially designed to amplify the B cell receptor (BCR) and T cell receptor (TCR) chains of the immune system with both inclusivity and quantitation in mind.
iRepertoire’s multiplexing solution
iRepertoire offers two different multiplex PCR approaches, both of which amplify all of the V(D)Js in a sample, including the highly variable CDR3 region. With arm-PCR (amplicon rescued multiplex PCR, patent 7,999,092) you can amplify the chain of your choosing from human or mouse samples with superior sensitivity so as to include all the diversity present in a sample, even for extremely rare clonotypes. With dam-PCR (dimer avoided multiplex PCR, patent pending), you can amplify any or all of the BCR and TCR chains in a much more quantitative manner than competing approaches.
Amplicon rescued multiplex PCR (arm-PCR)
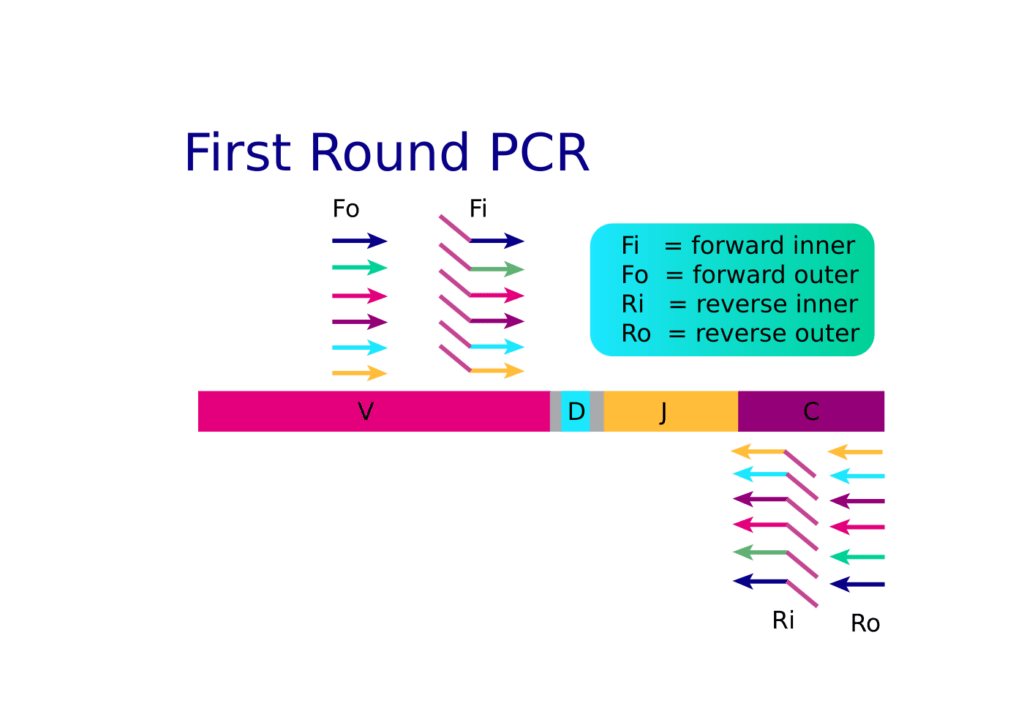
Arm-PCR uses a high concentration of hundreds of nested inside and outside gene-specific primers in the initial combined reverse transcription (RT) plus PCR round one. Both the reverse outside and inside primers can contribute to first strand synthesis during RT, which is especially important if there are any secondary RNA structures that make the inner primer binding site inaccessible during RT.
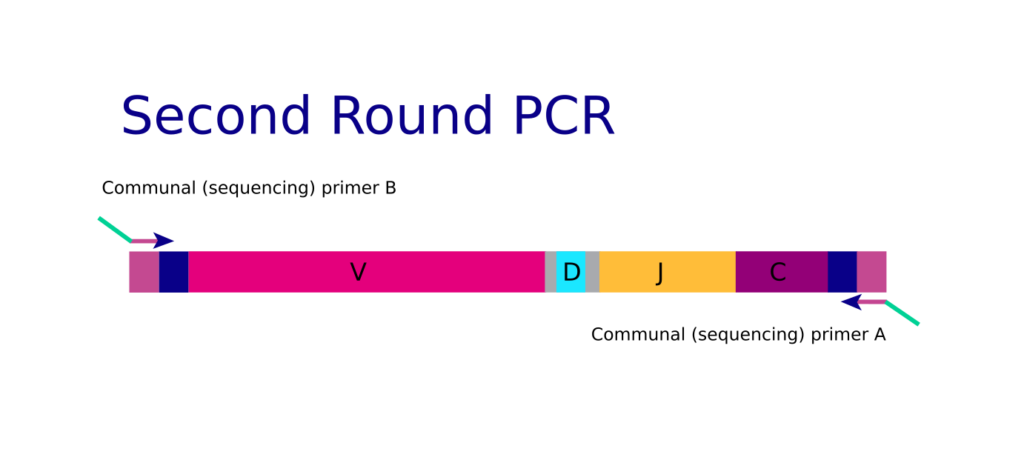
The outside primers help to improve the sensitivity of the reaction by increasing target template abundance for the inside primer to bind. Because the nested primer mix goes through many binding and extension cycles, arm-PCR is a great technological solution for rare clonotype discovery. When RT-PCR1 is complete, target amplicons are rescued, and PCR round two is performed using fresh enzymes. For PCR2, communal primers that recognize the shared tag sequence introduced during the first round of amplification are used for further amplification.
Dimer avoided multiplex PCR (dam-PCR)
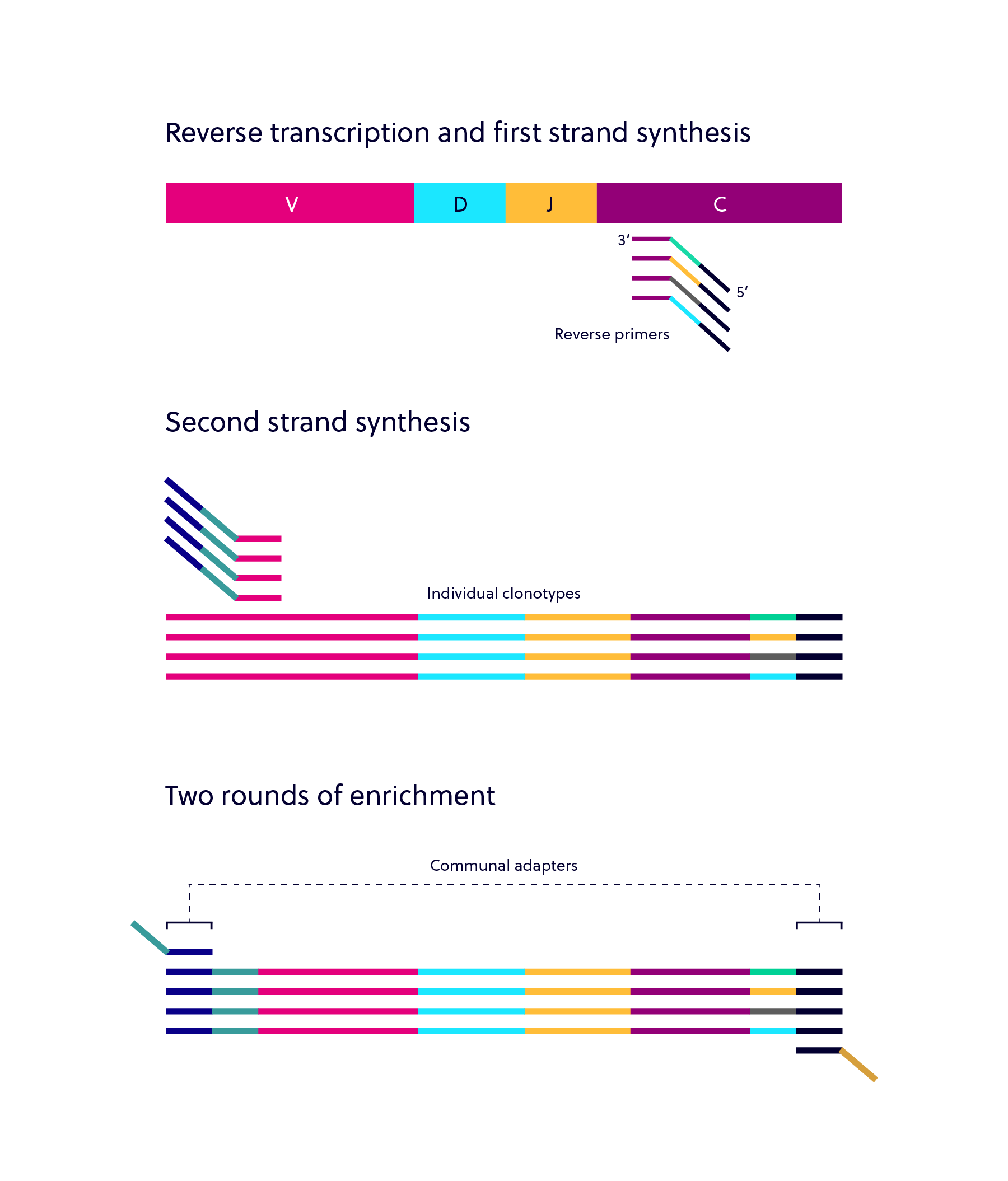
iRepertoire’s dam-PCR technology lets you select any combination of TCR and BCR chains for simultaneous amplification in one reaction. This is made possible by the unique single-cycle binding and extension steps and stringent clean up steps in-between, which omit harmful dimer formation. First, only the 3’ primer is added and one binding and extension step is performed. The unincorporated 3’ primers are washed away, and then the 5’ primers are added. After another single cycle binding and extension protocol, the 5’ primers are washed away. Primers that bind to the communal primer sites introduced in the first two steps are added for multi-cycle, exponential amplification.
Dam-PCR also allows for the inclusion of unique molecular indices (UMI) so that each strand of RNA can be tagged for direct quantification and both PCR and/or sequencing error removal. This increases confidence in the sequenced targets, and the quantification allows for investigation of interchain ratios in the immune adaptome. Thus, while arm-PCR provides an inclusive, semi-quantitative overview of the immune repertoire with respect to particular chains, dam-PCR provides a more quantitative look at the frequency of particular clonotypes of interest for any or all BCR and TCR chains.
iRepertoire has also developed an automation platform called iR-Flex to enable immune repertoire amplification by arm-PCR or dam-PCR in enclosed, contamination-free cassettes. Because dam-PCR requires extensive sample processing, dam-PCR is only offered via iR-Flex or through services.