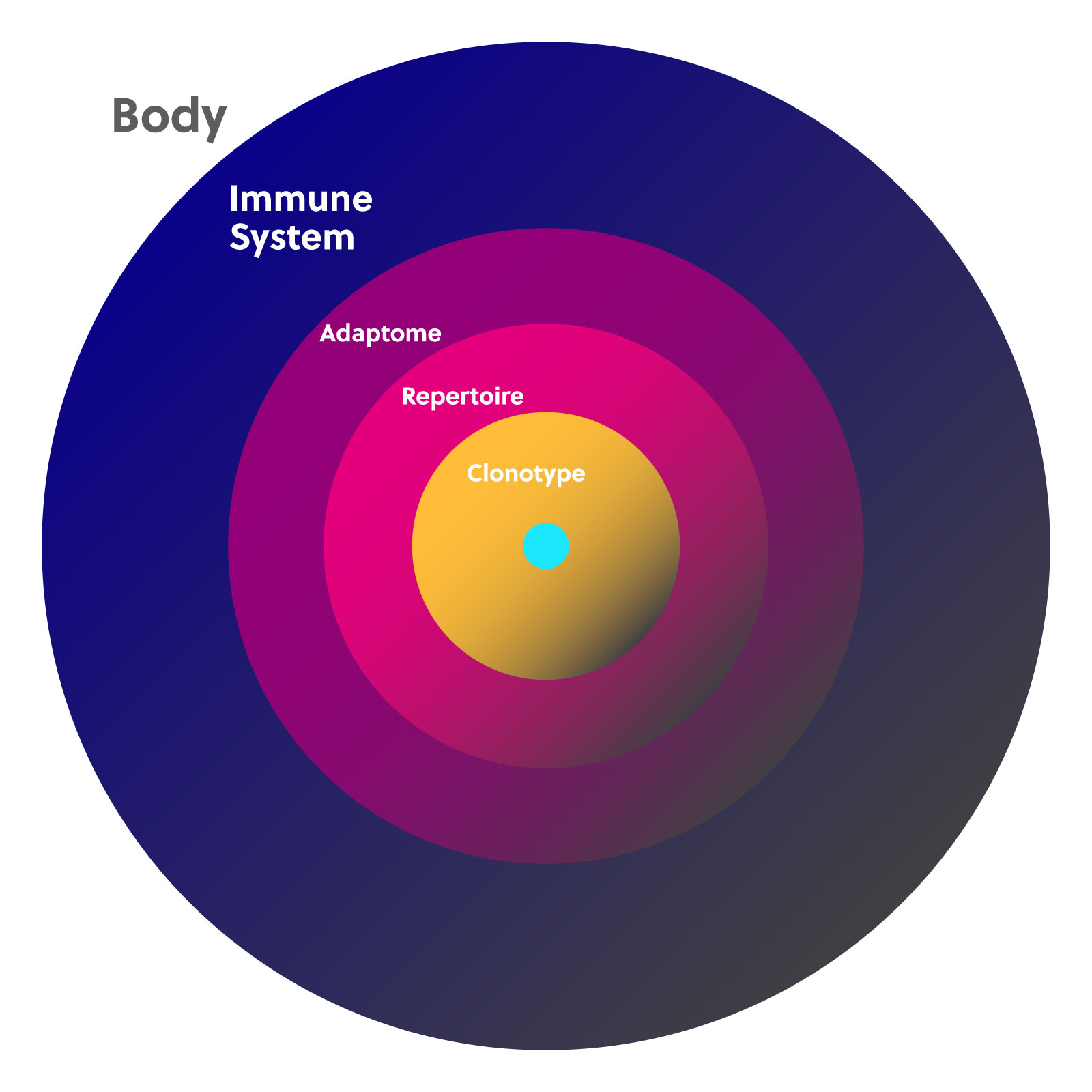
Defining the immune repertoire and adaptome
Each B and T cell produced by the body has only one receptor clonotype. Because the number of cells in the adaptive immune system is finite, if one clonotype expands, others will be displaced. The adaptive immune system is thus made up of a landscape of cells that fluctuate in number, which creates your personal immune diversity. This landscape is often described using the terms “immune repertoire” and “adaptome.”
An immune repertoire is the complete collection of individual clonotypes for any particular receptor chain, including the highly diverse CDR3 region. Studying the immune repertoire can reveal a lot about a person’s current health while also providing a historical record of their immune challenges and changes throughout their lifetime.
The adaptome encompasses all of the B and T cell receptor genes expressed in a given sample. This includes all of the T cell receptor (TCR) alpha, beta, delta, and gamma chains and all b cell receptor (BCR) heavy and light chains. The adaptome also takes into consideration the expression ratios of each chain in conjunction with each other.
For background information about BCR and TCRs, see our T cell and B cell overview page
Together, the immune repertoire and adaptome comprise the full landscape of T and B cells with all of their functional and structural diversity at any given point in time.
Methods for studying the repertoire and adaptome
Historically, B cells and T cells were studied by conducting a phenotypic analysis of the cell/bulk sample via immunostaining. Immunostaining can reveal receptor binding affinity to an extent, but it is insufficiently sensitive and not comprehensive enough to capture the full repertoire diversity quantitatively or qualitatively. Sequencing to identify the specific BCR and TCR genes that are being expressed can provide a much more complete picture of the immune repertoire and adaptome.
There are a number of approaches that can be used to sequence the immune repertoire. One option is to perform bulk RNASeq. RNAseq provides fairly comprehensive gene expression information, but its ability to capture numerous, complete VDJ sequences is lacking. This is because the VDJ region is rarely sequenced as one full read; rather, many different reads are stitched together post-sequencing via specialized software1. Alternatively, a technique called 5’RACE can be used to target TCR and BCR encoding transcripts more specifically. 5’RACE relies on reverse transcription beginning, ideally, in the C gene. Because reverse transcriptase has an extension limit (usually only a few kb), 5’RACE may be biased towards amplifying gene segments with short 5’UTRs.
Multiplex PCR (mPCR) is a promising method for repertoire analysis because it enables specific targeting of multiple desired products at once, but there are several challenges. Each primer needs to be validated, and pairs of primers amplifying different loci need to be compatible in the same reaction systems. That is, they need to perform under similar conditions and not amplify amongst themselves, which will produce primer dimers. Two multiplex PCR chemistries called arm-PCR and dam-PCR have been developed to address these challenges for sequencing the immune repertoire and adaptome specifically.
For more about arm-PCR and dam-PCR, visit our page on iRepertoire’s amplification technology
Immune diversity as a biomarker
Diversity in the immune repertoire correlates with a number of physiological states and diseases including cancer, autoimmunity, and aging. Diversity tends to decrease with disease progression and age. Immune repertoire diversity can thus potentially be diagnostic and prognostic.
For information about statistical methods for quantifying and analyzing immune diversity, visit our data analysis page
Several recent studies have demonstrated how immune repertoire diversity correlates with disease state. Last year, researchers at the Chinese Academy of Medical Sciences showed that TCR-gamma CDR3 diversity is significantly reduced in patients with lung cancer carcinoma2. Conversely, researchers at the University of Alabama found that ulcerative colitis (UC) and Crohn’s disease (CD) patients have significantly more diverse TCR-beta CDR3s3. Further there were disease-specific clonotype expansions that were not shared between UC and CD patients.
Medical research applications of immune repertoire sequencing
An understanding of the particular lymphocytes that proliferate during disease could help develop vaccines or treatments. For instance, sequencing has been used to identify the specific VJ rearrangement patterns of gamma/delta T cells that proliferate following tuberculosis (TB) infection4. The only currently available vaccine for TB is not particularly efficacious in adults, so it is critical to understand the body’s natural mechanism of protection.
The prognosis of related diseases may result in convergent evolutions of the same or similar clonotypes in different patients. Identifying two unique CDR3’s, called linklets, that co-occur in patients with the same disease can be especially powerful. Repertoire analysis could thus be diagnostic or prognostic. Research by the NIH earlier this year also suggests that there may be a specific CDR3 signature associated with multiple sclerosis (MS)5. There is currently no clear diagnostic indicator for MS. Similarly, hepatitis patients with a favorable disease outcome tended to share particular TCR-beta chains6. These clonotypes could thus serve as a biomarker for favorable prognosis.
Finally, repertoire analysis could be used to track or predict response to treatment. Treatment response in patients with end-stage renal disease has already been shown to correlate with V and J gene usage in TCR-beta chains7. Repertoire analysis could also be used to study and track tumor-infiltrating lymphocytes and clonotypes associated with the cancer microenvironment before and after treatment.
For more case studies of TCR and BCR composition, see our page on sequencing the immune repertoire
References
1. Bolotin, D.A. et al. Antigen receptor repertoire profiling from RNA-seq data. Nat Biotechnol. 35(10), 908-911 (2017).
2. Chen, H. et al. Profiling the pattern of the human T-cell receptor γδ complementary determinant region 3 repertoire in patients with lung carcinoma via high-throughput sequencing analysis. Cell. Mol. Immunol. 16, 250–259 (2019).
3. Wu, J. et al. Expanded TCRβ CDR3 clonotypes distinguish Crohn’s disease and ulcerative colitis patients. Mucosal Immunol. 11, 1487–1495 (2018).
4. Cheng, C. et al. Next generation sequencing reveals changes of the γδ T cell receptor repertoires in patients with pulmonary tuberculosis. Sci. Rep.8, 1–13 (2018).
5. Sousa, A. de P. A. et al. Comprehensive Analysis of TCR-β Repertoire in Patients with Neurological Immune-mediated Disorders. Sci. Rep. 9, 1–10 (2019).
6. Jiang, Q. et al. Patient-shared TCRβ-CDR3 clonotypes correlate with favorable prognosis in chronic hepatitis B. Eur. J. Immunol. 48, 1539–1549 (2018).
7. Wong, H. S.-C. et al. V-J combinations of T-cell receptor predict responses to erythropoietin in end-stage renal disease patients. J. Biomed. Sci. 24, 43 (2017).