Capturing the full clonal diversity of the immune repertoire via sequencing can reveal important information about the pathology and prognosis of disease. Immune repertoire sequencing for patients with different diseases may show quantitative or qualitative differences from healthy controls in terms of repertoire composition and diversity.
Qualitative changes may present as increased sharing of disease-specific CDR3s in T or B cells. Quantitative changes may manifest as increases and decreases in repertoire diversity. Capturing both qualitative and quantitative changes requires an immune repertoire sequencing approach that is both sensitive (so as to include all CDR3s) and unbiased (to allow for relative quantitation of all CDR3s).
iRepertoire’s immune repertoire sequencing approach is:
- Highly sensitive for inclusive analysis
- Unbiased to allow for quantification
- Robust to cover critical antigen recognition regions
- Uniquely tailored to provide specific information
For examples of how immune repertoire sequencing applies to health, see the Learning Center Article “Why sequence the immune repertoire?”
Quantitative and inclusive immune repertoire analysis
While there are hundreds of thousands of different clonotypes in one person’s immune system, only a small number of clonotypes make up the bulk of the immune repertoire. Sampling the CDR3 clonotypes in a single person would thus produce a distribution like the one shown in the graph below.
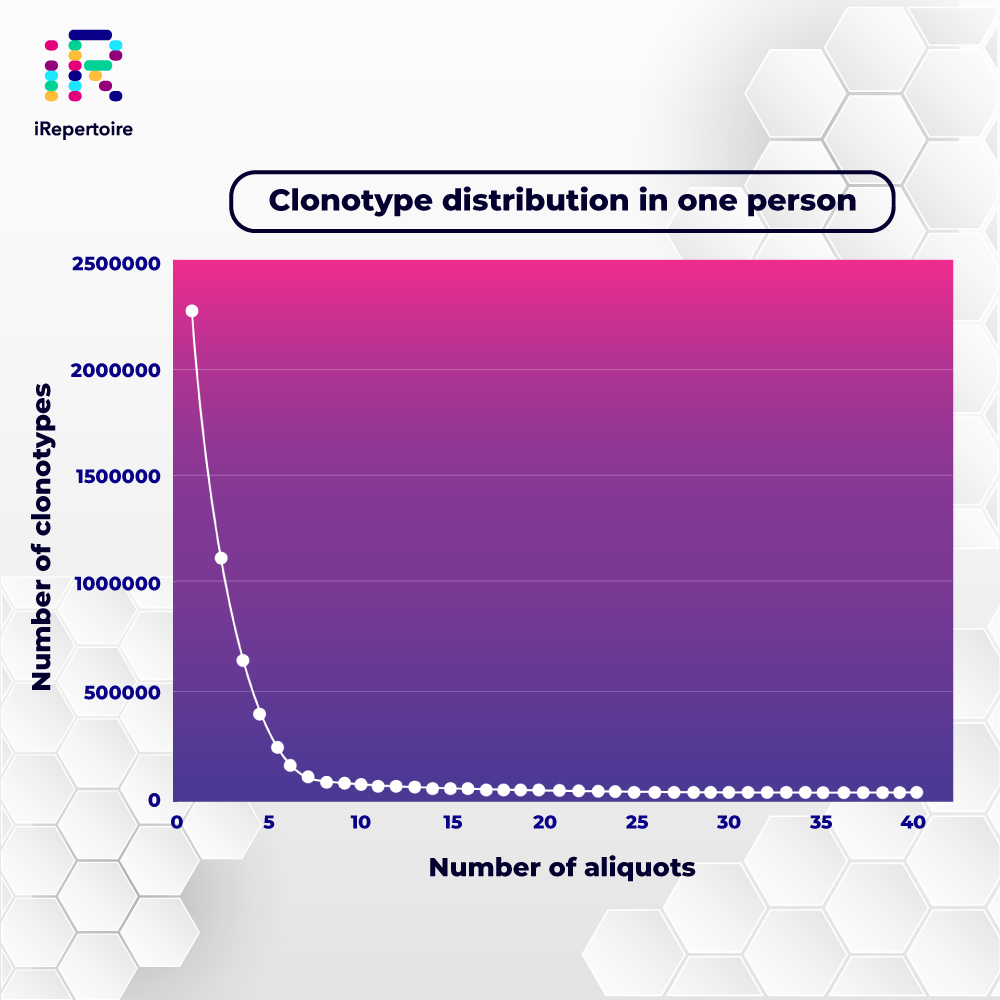
This long-tailed distribution results in a low signal-to-noise ratio, as the more frequent CDR3s will be overrepresented in an immune sequencing library. To effectively capture the full breadth and depth of the immune repertoire, the sequencing methods must, therefore, be inclusive and quantitative.
Information about the absolute quantity of particular clonotypes can be difficult to attain because amplification is required to prepare libraries for sequencing. The amplification process is biased towards abundant and small amplicons, which can cause less frequent, rare amplicons to be underrepresented in the final library. iRepertoire developed two different multiplexed PCR methods to increase rare clonotype discovery and reduce these biases in their immune repertoire sequencing technologies.
These technologies are described in depth in our amplification technology post.
Deciding what to sequence
An experimenter wishing to sequence the immune repertoire will need to determine what region(s) of the T cell and/or B cell receptor (TCR and BCR) genes to target, whether to use DNA or RNA as starting material, and whether to sequence bulk tissue or single cells, all of which depend on the goals of the study.
Choosing what region to sequence:
The regions of TCRs and BCRs that play the most important role in antigen recognition are also the most variable. These variable regions are constructed from a random shuffling of gene segments, and, in the case of BCRs, somatic mutations. The most variable portion of the variable region is the CDR3 region. It is, therefore, critically important to target the CDR3 region when immune repertoire sequencing. All of iRepertoire’s primer systems cover the highly variable CDR3 region.
Sequencing the full variable region can provide more information, but that extra information comes at the cost of increased sequencing reads or decreased sequencing coverage. For more about sequencing coverage, see our post about choosing read depth.
Choosing the starting material:
While both RNA and DNA sequencing can provide a picture of the immune repertoire, only RNA sequencing will reflect the library of TCR and BCR sequences that are actually expressed (for more information, see our post on template selection).
Choosing bulk tissue or single cell sequencing:
Finally, bulk sequencing is required to analyze immune repertoire diversity, but information about the cognate pairing of BCR/TCR chains within a given B or T cell is lost in bulk sequencing. Researchers that wish to examine the relationships between paired chains, in particular clonotypes, should perform single cell sequencing. For more information about how to sequence single cells, see our post on cell sorting.